At the beginning of the pandemic, while I was working at Las Condes Hospital, a project was developed together with Amazon Web Services Chile for the detection of COVID-19 pneumonia patterns in chest X-rays. Using weighted cost functions for unbalanced data, transfer learning and augmentation. An accuracy of 0.92 was achieved with only four thousand images. The project was a success in every sense, used in many medical centers we receive a lot of positive feedback, and get a lot of attention from the main media in the country, including news, radio, and television.
Introduction

Project in AI algorithm for the automatic detection of COVID-19 pneumonia patterns in frontal chest X-ray images to combat the pandemic. Currently, at the time of writing this document, the main disadvantage of the official PCR test is that it can take many hours to obtain a result, and even several days, which is a big problem with the current collapse of the health system and the increasing need for specialized laboratories with their limited supplies problems (at least for now there are not present in many places across the country). Instead, our system is instantaneous, easily accessible and uses minimal resources in the number of possible tests. Also, it can be used as a complement to PCR, which also has a considerable failure rate, always considering the pandemic context and accompanied by the correct symptoms of the disease. In short, it provides immediate help in the absence of PCR or the shortage of expert radiologists.
Data
The database is made up of three sources:
| SOURCE | TYPE | NUMBER OF SAMPLES |
| https://www.kaggle.com/c/rsna-pneumonia-detection-challenge | WITHOUT COVID | 3014 |
| INTERNAL CLC | WITH COVID | 573 |
| https://github.com/ieee8023/covid-chestxray-dataset | WITH COVID | 218 |
The main objective of the system was to determine the presence of the disease in the early stages, when the person does not present great damage in the lungs. Expert radiologists cleaned images of patients with the disease but no visible patterns of COVID pneumonia. But it is also important that the system does not confuse any spot in the lung with COVID, for this reason most of the non-COVID images used correspond to other pathologies and not only to healthy patients.
The images were reviewed, validated and selected by expert radiologists, given that not all patients present distinguishable patterns of COVID on their radiography. What in principle could cause errors and was one of the main problems at the beginning of the project, because before this correction, false positives were very high (totally healthy patients falsely diagnosed with the disease).
It should be noted that the first set are external images that come from the RSNA and are validated for a Kaggle competition from before the pandemic. All correspond to patients without COVID, who are certified for competition in the highest current standards for the generation of AI models.
Order
Being a small data, three sets were formed completely randomly. One used for model training, one for validation to calibrate hyperparameters and another for final testing, which was only used once the project was finished. The validation and test set where all taken from the same source, this to avoid that the model was recognizing the source instead of the COVID patterns.
| positive | negative | total | |
| train set | 741 | 2964 | 3705 |
| validation set | 50 | 50 | 100 |
| test set | 33 | 33 | 66 |
| total | 824 | 3047 | 3871 |
In the training set we have four times fewer COVID cases. Different weights must be entered for each category in the cost function to be optimized in training. Note that, given computational constraints, the data must be passed in batches. These batches must be large enough (more than 100 images each), such that they contain enough COVID images in each step of the training.
To assess the model’s accuracy, it’s crucial to consider that the validation and test sets are balanced. This balance is essential due to the limited data availability and the significant shifts in data distribution across various hospitals, geographical locations, and over time. The fluctuation in the number of positive and negative cases is particularly pronounced as the pandemic evolves, with cases potentially decreasing substantially during extended lockdowns. Additionally, the distribution disparity is notable in diverse localities where the model is applied. The a priori probability undergoes substantial changes, exerting a significant influence on the final metrics and emphasizing the need for a nuanced interpretation of the results.
Preprocessing
U-Net
Due to date and other origin annotations present on the images, we decided to remove all annotations of the images by using an automatic segmentation system to avoid bias. A U-Net type AI architecture (currently the most modern for this type of task), was trained to perform automatic segmentation of the lungs in the images before classifying it for disease detection (see figure 1).
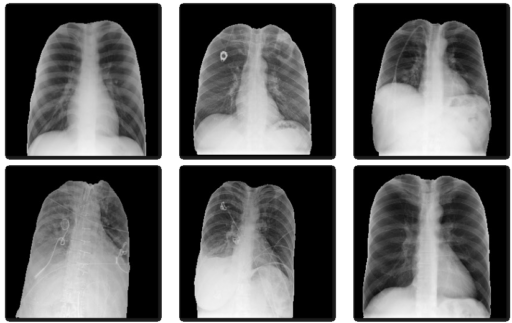
Augmentation
The data is increased by applying different transformations randomly as shown in the following table.
| TYPE OF TRANSFORM | RANGE |
| Horizontal deformation | 0.05 |
| Rotation range | 20 |
| Zoom range | 0.2 |
| Width change range | 0.2 |
| Height change range | 0.2 |
| Brightness change | 0.1 to 1.9 |
| Horizontal flip | True |
This gives more variability to the data and helps the model to generalize so that it responds better to new data and with different distributions, with the aim of being more robust and reliable. See figure 2.
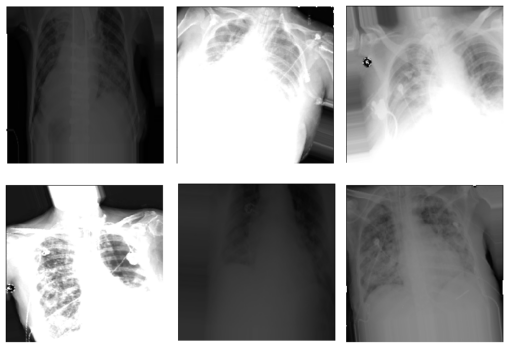
This is very important, for example, we have to consider that even for the same machine it can have very different configurations, also the machines can vary a lot for each center (you can think of it as different normal cell phone cameras, with their own configurations). Also, some person could take a simple picture with his cell phone of the screen where the original X-Ray really is, and alter the intensity or angle of the picture.
Classification model
Given that relatively small data sets are available, it was decided to use transfer learning of a classical VGG16 model pretrained in the first convolutional layers with fixed weights obtained on ImageNet images. This set contains a thousand categories of images of all kinds, not from the medical area, so in the first fixed convolutional layers the model contains the filters for the relevant characteristics typical of any image (for example, edge detection).
For the unfrozen layers of the model it is necessary to change the last dense layers to be of two categories. The unfixed layers are then retrained for a few epochs on a different problem. This time a problem from the medical area is used, with chest x-rays for the detection of pneumonia with over twelve thousand images of data from a Kaggle competition (with images very similar to the final problem to be solved, but where there are many more images). Also, since COVID can cause a type of pneumonia, the problems are highly related and it helps the system a lot.
Finally, you train on the final problem. During training, some neurons are randomly removed (given the scarcity of data, to avoid overfitting the training set and poor generalization). In addition, given the class imbalance, a weighted cost function is used to optimize during this last training.
Results
The values over the validation set, even distorting the images, are shown below:
| METRIC | VALUE |
| True positive | 48 |
| False positive | 5 |
| True negative | 45 |
| False negative | 2 |
| Accuracy | 0.93 |
It is emphasized again that since it is a medical exam and the a priori probability of COVID changes a lot, in both time and region, and it is very hard estimate.
When testing the model in practice on the test set once the project is finished, with only images from our hospital to ensure that the system does not have any type of bias due to the origin of the data. The system gave a final accuracy of 0.92 over the 66 images (with a table almost identical to that of the validation set).
Tools: Python, Git, NumPy, OpenCV, TensorFlow, Keras, CUDA/GPU, AWS SageMaker, AWS Lambda, AWS SNS, 3D Slicer, Matplotlib, Scikit-Image.
